製品・サービス情報
|
|
Arraystar社 次世代シークエンス(NGS)
R-loop profiling 受託解析サービス |
|
 |
Arraystar社(アメリカ)との業務提携による 次世代シークエンシング(次世代シーケンシング/NGS)サービスでは、サンプルからデータへのソリューションを提供します。R-loop
profiling では、ゲノムにおける LncRNA/mRNA の組織化された R-loop の分布をプロファイルします。
本サービスのバイオインフォマティクス解析では、生物学的および機能的な知見が得られます。R-loop profiling では、S9.6 抗体を用いて、R-loop
を特異的に免疫沈降させます。R-loop の RNA 鎖を配列決定し、解析します。 |
 特長 特長
|
| ◆ |
強力なプロファイリング:ゲノム上の遺伝子制御における新たな担い手としての R-loop 研究 |
| ◆ |
ストランド特異性: どの LncRNA または mRNA が R-loop を形成するかを特定する。 |
| ◆ |
高い信頼性: 確立された最適な実験手順による最良の結果 |
| ◆ |
柔軟性: DRIPc-Seq は、参照ゲノムを持つ任意の種に対して実行 |
| ◆ |
厳格な品質: ポジティブおよびネガティブコントロールによる DRIPc-Seq ライブラリの品質の保証 |
| ◆ |
視覚化: 豊富なアノテーション、ゲノムブラウザートラック、および論文に利用可能な図の提供 |
◆R-loop による制御
|
R-loopは、転写された RNA 鎖と鋳型 DNA 鎖が塩基対を形成した RNA:DNAハイブリッドと、非鋳型一本鎖 DNA から成る三本鎖構造です(図1)[1]。R-loop
は広く分布しており、哺乳類ゲノムの 5%に存在しています[2, 3]。R-loopは多くの場合、プロモーターの CpG アイランドや転写停止部位に存在します。R-loop
の形成には、高GCスキュー(TSS 非テンプレート鎖の下流において C よりも G が豊富)、G-quadruplex、DNA ギャップ、DNA/RNA
修飾が寄与しています[4]。R-loopは、遺伝子制御、DNA複製、DNA/ヒストン修飾など、生物学的に重要な機能を持っています。
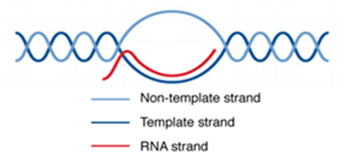
図1. R-loop の構造[5]。 |
【R-loop による DNA メチル化と mRNA の転写の制御】
|
通常、BAMBI(TGFb のネガティブレギュレーター)遺伝子プロモーターの R-loop は転写を促進する。しかし、筋萎縮性側索硬化症(ALS4)では、senataxin
の変異により R-loop が減少し、BAMBIプロモーターでのDNAメチル化が増加し、BAMBI転写抑制、TGFb シグナル伝達上昇、ALS
進行につながります(図2)[6]。
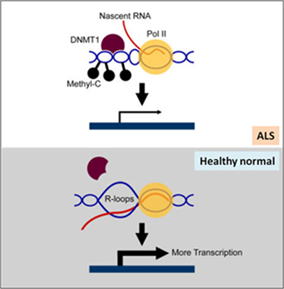
図2. 健康な正常細胞では、BAMBI プロモーターのR-loopが転写を促進します。
ALS細胞では、Senataxin 変異がプロモーターでの R-loop 形成およびBAMBI 転写を抑制します。 |
|
【アンチセンス lncRNA R-loop は、mRNA 転写に影響を与える
|
R-loop は、アンチセンスRNA鎖とループ内のDNAとの間に形成される。TCF21 は多くの癌の腫瘍抑制因子です。TARID(脱メチル化を誘導する
TCF21 アンチセンスRNA)は、TCF21 遺伝子の一対一のアンチセンス lncRNA であり、プロモーター領域に R-loop を形成しています(図3)。この
R-loop は、GADD45a によって認識され、脱メチル化酵素 TET1 をリクルートして DNA メチル化を除去し、TCF21 mRNA
転写を増加させ、細胞周期を制御します[7]。
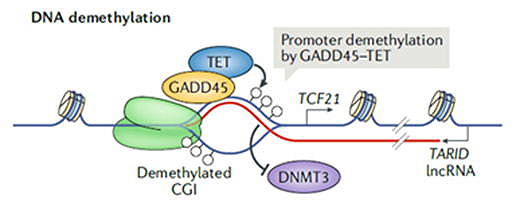
図3. アンチセンス lncRNATARID は、R-loop を形成し、TCF21 プロモーターの脱メチル化と TCF21 mRNA の転写を制御する[4]。 |
|
【関連レビュー】
|
| 1. |
Hamperl S. and Cimprich KA. (2014) The contribution
of co-transcriptional RNA:DNA hybrid structures to DNA damage and genome
instability. DNA Repair (Amst) 19:84-94 [PMID:24746923] |
| 2. |
Sanz LA. et al (2016) Prevalent, Dynamic, and Conserved R-Loop Structures
Associate with Specific Epigenomic Signatures in Mammals. Mol Cell 63(1):167-78
[PMID:27373332]
|
| 3. |
Li M. and Klungland A. (2020) Modifications and interactions at the
R-loop. DNA Repair (Amst) 96:102958 [PMID:32961406]
|
| 4. |
Niehrs C. and Luke B. (2020) Regulatory R-loops as facilitators of gene
expression and genome stability. Nat Rev Mol Cell Biol 21(3):167-178
[PMID:32005969]
|
| 5. |
Hegazy YA. et al (2020) The balancing act of R-loop biology: The good, the
bad, and the ugly. J Biol Chem 295(4):905-913 [PMID:31843970]
|
| 6. |
Grunseich C. et al (2018) Senataxin Mutation Reveals How R-Loops Promote
Transcription by Blocking DNA Methylation at Gene Promoters. Mol Cell
69(3):426-437.e7 [PMID:29395064]
|
| 7. |
Arab K. et al (2019) GADD45A binds R-loops and recruits TET1 to CpG island
promoters. Nat Genet 51(2):217-223 [PMID:30617255]
|
|
|
|
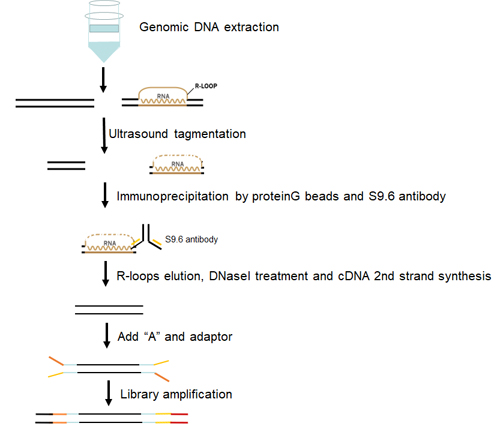
 解析の流れ 解析の流れ
|
本サービスは、gDNA~データ解析までのフルパッケージとなっています。
 |
|
【解析ワークフロー】
|
1. |
gDNA サンプルの受け取り |
| 2. |
超音波処理 |
| 3. |
S9.6 抗体を用いた DNA-RNA 二本鎖免疫沈降 |
| 4. |
ストランド特異性ライブラリー調製 |
| 5. |
クラスターステーションでのクラスター形成 |
| 6. |
Illumina プラットフォームによるシークエンシング |
| 7. |
データ抽出と解析およびまとめ |
|
【バイオインフォマティクス解析】
|
本サービスのバイオインフォマティクス解析では、以下のデータを提供します。
| ・ |
サンプル QC 結果(gDNA サンプル QC および DRIPc QC) |
| ・ |
シークエンスデータ(Clean reads および mapped reads) |
| ・ |
データ解析:
| - |
シークエンスデータのQC |
| - |
ゲノム参照データベースへのリードマッピングとアライメント |
| - |
R-loop ピーク解析:R-loop RNA 種類(lncRNA または mRNA)、R-loop ピーク分類、gene feature 上の分布とエンリッチメントおよび遺伝子本体内での分布 |
| - |
R-loop ピークの変動解析(fold change≧2 and p-values≦0.05) |
| - |
R-loop ピークのボルケーノプロット |
| - |
有意差のある R-loop ピークを含む遺伝子の GO/Pathway 解析 |
|
| ・ |
プロジェクトサマリーレポート |
|
【データ例】
|
・ピークコールとアノテーション
|
統計的に優位な DRIP 濃縮領域(R-loop ピーク)は、MACS2により p 値の閾値を 0.001 としてコールされます。
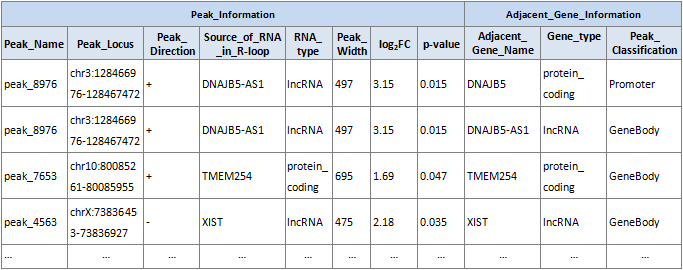
|
・R-loop ピークの分類
|
R-loop ピークは、UCSC RefSeq アノテーションに基づく直近の遺伝子特徴(プロモーター、遺伝子本体、ターミネーター、遺伝子間)により分類されます(図4)。R-loop
の分類分布のサマリー統計が提供されます。
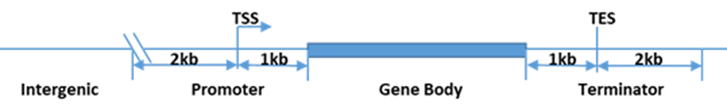
図1. 4種類の遺伝子特徴における R-loop ピーク分類。 |
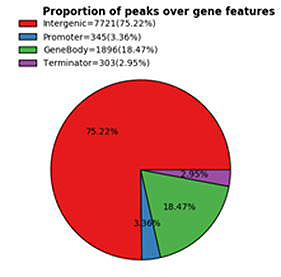
図2. 遺伝子特徴分類における R-loop ピークの割合。 |
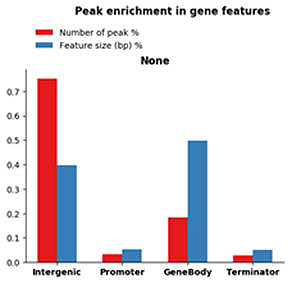
図3. 遺伝子特徴における R-loop ピークの濃縮度と、機能サイズに対して正規化されたランダム偶数分布の比較 |
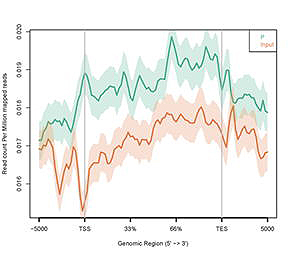
図4. 遺伝子本体上の R-loop ピーク分布。
X軸は転写開始点(TSS)と転写終了点(TES)間の遺伝子本体
(遺伝子本体の長さを100%に標準化)を表しています。
Y軸はマッピングされた 100万リードあたりのリードカウントを示します。 |
|
|
|
 サンプル条件 サンプル条件
|
| 品質条件 |
詳細 |
| 対象生物種 |
Human/Mouse/Rat(その他生物種については、お問合せください。) |
| サンプルタイプ |
gDNA: フェノール-クロロホルムで抽出してください。
※R-loop 構造の破壊を避けるため、、抽出作業中にボルテックスをしないでください。 |
| 必要量 |
>10μg |
| 濃度(Nanodropに基づく) |
>20ng/μL |
| 純度(Nanodropに基づく) |
O.D.260/280:1.7
O.D.260/230:1.8 |
| DNA Integrity |
・アガロースゲル電気泳動: 高分子DNAのバンドがはっきりと確認できること。 |
| ※ |
DNase、RNase フリーの 1.5ml または 2ml マイクロチューブを使用してください(スクリューキャップ式を推奨) |
| ※ |
ヌクレアーゼフリーの分子生物学グレードの水に溶解し、-80℃で保存してください。 |
| ※ |
UV スペクトルベースの濃度測定法では、不正確になる場合がありますので、蛍光ベースでの測定を強く推奨します。 |
| ※ |
UV スペクトルベースで測定した場合、上記よりも多いサンプルをご準備ください。 |
|
 価格 価格
|
| サービス名 |
税別価格 |
カタログ# |
R-loop profiling 受託解析サービス
(including IP+input) |
¥361,000 |
F-AS-DRIPcseq-[サンプル数] |
| ※ |
1サンプルの解析料金です。海外提携先への送料が別途必要になります。 |
|
 ご注意事項(必ずご確認ください) ご注意事項(必ずご確認ください)
|
| 1. |
解析サービスに関して
本サービスの解析は、弊社の海外業務提携先(Arraystar社)にて実施されます。 |
| 2. |
解析に利用する製品に関して
本サービスの仕様は、事前の予告なく変更される場合があります。 |
| 3. |
お見積りに関して
正式なお見積書は、直接販売の場合には直接ご提出いたしますが、代理店経由の場合は代理店よりご提出させて頂きます。 |
| 4. |
サンプルの送付および取り扱いに関して
| 4.1 |
サンプルのご発送は、必ず、サンプル送付方法およびご注意点(PDF)の内容に従ってください。送付の際は、チューブをパラフィルムで覆い、キャップの緩みや中身の漏れがおきない様にしてください。サンプルの入ったチューブには、油性マジックでサンプル名を明記し、ビニールバッグ等に入れてください。DNAサンプルは、アイスバックなどとともに梱包し、クール便でお送りください。ただし、DNAが冷凍状態で保存されている場合は、砕いたドライアイスとともに梱包して、冷凍便でお送りください。 |
| 4.2 |
サンプルは、なるべく休日をはさまない様に、弊社営業時間内(土日祝日、年末年始等を除く、平日9時~18時)に到着する様にご発送ください。なお、ご発送時の送料はお客様負担となります。着払いでは受け取りできませんので、あらかじめご注意ください。 |
| 4.3 |
サンプル送付の際は、受託解析サービスQCシートおよび免責事項同意書を同封してください。これらが同封されていない、または記入漏れがある場合、解析サービスに着手できませんので、ご注意ください。 |
| 4.4 |
ご送付いただいた DNA の品質チェックの結果、解析に必要な量が不足または低品質であった場合、サンプルの再送をお願いする場合があります。なお、再度サンプルをお送り頂く場合、業務提携先への再送料金をご負担いただきます。 |
| 4.5 |
弊社に起因しない(国内および国際輸送時など)サンプル喪失等に対する補償・保険制度はありませんので、貴重なサンプルをご提供頂く際はご注意ください。 |
| 4.6 |
お預かりできるサンプルは、BSL2(バイオセーフティーレベル2)までのものに限られます。感染性が著しく高いサンプル(HIV、HCV、HBVウイルス)に感染していることが確認されている患者由来の検体など)は、お預かりできませんので、予めご了承ください。ヒト臨床サンプルの場合、インフォームドコンセントを得てからご提供ください。 |
| 4.7 |
ご提供頂いたサンプルの返却は致しておりません。(サンプルは解析終了時に破棄されます。) |
| 4.8 |
本確認事項を満たさない事で別途費用が発生した場合、お客様に費用のご負担をお願いすることがあります。 |
| 4.9 |
サンプル・業務等から生じた知的財産権・工業所有権・安全性・インフォームドコンセント等の問題については、弊社は一切の責任を負わないものとします。 |
|
| 4. |
キャンセルに関して
本サービスは、海外での解析という性質上、サンプル受領後のキャンセルはお引き受けできません。やむを得ない理由でキャンセルされる場合、それまでの工程に応じた料金を請求いたします。
|
| 5. |
納品物に関して
納品物は、代理店経由でのお取引の場合、基本的に代理店を介してお送りします。
納品データのバックアップは、必ずお客様にてお取りください。弊社でのデータ保管期間は、発送日から90日間です。弊社での保管期間を経過したデータは、破棄されます。 |
|
本ページは、Arraystar社のホームページに掲載されている情報や画像を一部引用しています。
|